|
|
|
Research Research Areas Research Areas
Scientific Mission
Our Soft Matter Physics group focuses on a quantitative understanding
of biological cell phenotype and function from a complex, emerging materials
perspective encompassing all length scales from molecules to tissues, which
places our group in a pioneering and leading position in the novel area
Physics of Cancer.
Cancer cell unjamming and collective motility
The mechanics and physics of cancer are not quite fully understood. It is not yet clear why tumors
are often hard. Which roles play the dense cell clusters, and which the extracellular matrix?
Until recently, it was not even clear how and if cells in tumors move!
Our lab has considerably progressed the whole field (see list of publication below) on following questions:
- What is the state of matter of a living tissue? We show that real tissues can effectively make a phase transition between fluid-like and solid-like.
- How do cells move in dense tissues in the body? We show that cells have to deform and elongate to squeeze through the dense tissues.
Their motility state is visible in static images.
- What is the structure of dense tissues made of cells? We find out how single-cell properties (e.g. cell-cell adhesion, the glue between cells) are
connected to emerging tissue properties, such as surface tension, tissue cohesion, and the structure of tissues in general.
- Can we use this knowledge to improve diagnosis?
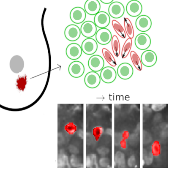
 |
Elongated cells are more motile and unjam tissues.
Left: Motion of a MDA-MB-436 cell in a densely packed spheroid.
For better visibility, the cell nucleus of interest is marked in red.
Right: Confocal image section through a MDA-MB-436 spheroid.
|
If you want to know more on one of our key insights, read this short synopsis:
Elongated Cells May Unjam Cancers
Main techniques:
- fluorescence live microscopy
- 3D cell culture
- 3D cell tracking
- Image analysis of cells and nuclei, their shapes
Key publications:
Cell jamming in real cancer tissues:
Grosser et al, Cell and Nucleus Shape as an Indicator of Tissue Fluidity in Carcinoma Phys. Rev. X (2021)
DOI: 10.1103/PhysRevX.11.011033
How cell-cell adhesion and the extracellular matrix influence if a tumor can metastasize:
Ilina et al, Cell-cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion Nat. Cell. Biol. (2020)
DOI:10.1038/s41556-020-0552-6
Cancer is (in some aspects) inverse to embryogenesis:
Blauth et al, Jamming in Embryogenesis and Cancer Progression Front. Phys. (2021)
DOI: 10.3389/fphy.2021.666709
Mechanical characterization of healthy epithelial and tumor spheroids
The mechanical behavior of biological tissue should be highly based on their composite single cell properties, e.g., viscoelasticity, cellular adhesion, contractility,
motility, proliferation, etc. Nonetheless, recent studies have illustrated in complex tissues that tissue mechanics is impacted by collective cell behavior, such as cell
unjamming and extended differential adhesion.
To link the properties of cancerous single cells and our primary tumor tissue, our lab are able to use Atomic Force Microscope (AFM) to characterize the behavior of healthy
epithelial and tumor cells in 3D spheroids.
The questions we are looking for in this field include (but not limited to):
- Do our healthy epithelial and tumor cells behave similarly in 2D monolayer and 3D spheorids?
- How does tissue surface tension play in a role?
- How do the jamming and unjaming correlates the behavior of cells in cell clusters?
- By modulating the cell cytoskeleton, determining its role in cell clusters.
Further investigation will be done to study the correlation between the cell nuclei shape and its properties (etc. viscoelasticity, motility, contractility)
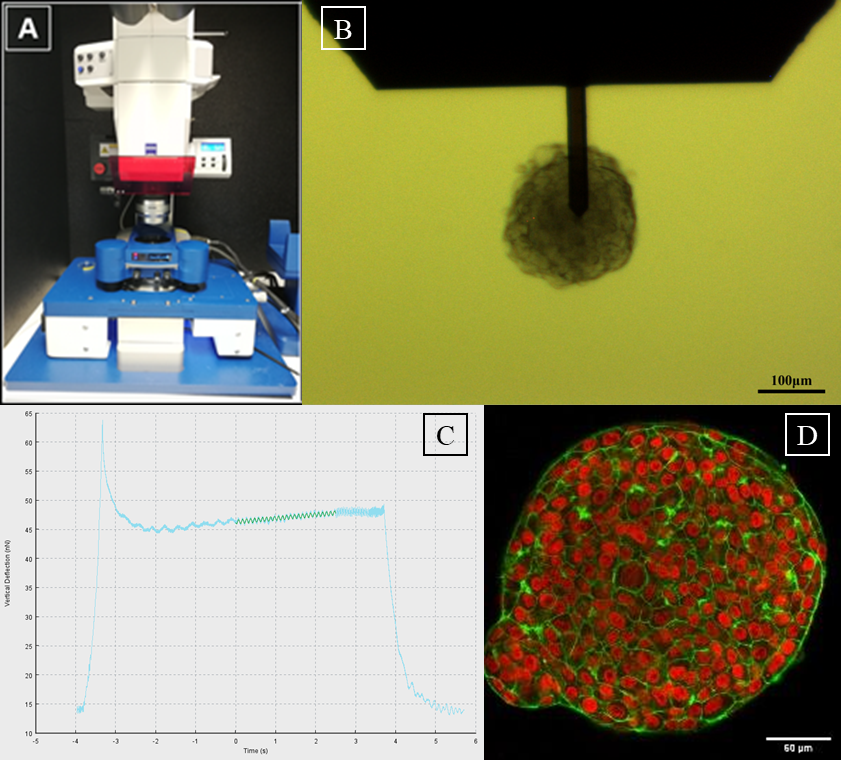 |
A: AFM setup, consisting of a Zeiss AxioZoom.V16 fluorescence microscope
equipped with a JPK Hybridstage™
B: MCF-10A spheroid phase contrast image
under AFM cantilever (TL-FM, Nanosensors, 28±7.5µm cantilever width) during
measurements (scale bar, 100µm)
C: Data processing image of sinusodal curve
fit using JPK data processing program, to evaluate G'/G'' of the corresponding
cell cluster
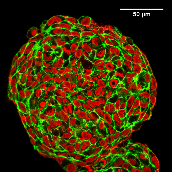
D: Equatorial confocal sections of MCF-10A spheroids. Cell nuclei
are marked in red. Actin, which assembles to a cortex beneath cell membranes,
is marked in green (scale bar, 50 µm), global contrast adjusted. |
Main techniques:
- atomic force microscopy
- 3D cell culture
- live fluorescence staining and immunostaining
- phase contrast and fluorescence microscopy
Key publications:
E. Warmt et al, Differences in cortical contractility between healthy epithelial and cancerous mesenchymal breast cells New Journal of Physics (2021)
DOI: 10.1088/1367-2630/ac254e
Grosser et al, Cell and Nucleus Shape as an Indicator of Tissue Fluidity in Carcinoma Phys. Rev. X (2021)
DOI: 10.1103/PhysRevX.11.011033
Collaborators:
Dr. Thomas Fuhs (former PWM group leader), TU Freiberg, Germany
Prof. Dr. Max Bi, Northeastern University, Boston, USA
Dr. Amit Das, Northeastern University, Boston, USA
Mechanical characterization of primary tissue
Tumorgenesis is associated with drastic tissue changes and thus an alteration in inherent tissue mechanics. So far, these have played no significant role in diagnosis of tumor malignancy.
Biophysicists already performed various in vitro studies and found that single cancer cells are softer compared to normal cells, whereas tumor tissues often appear more solid than their surroundings.
We apply a multiscale-approach towards a holistic tissue characterisation from bulk properties down to single cells. We analyze cancerous and non-cancerous human primary tissue samples of breast and
cervix are measured by novel Tabletop-Magnetic Resonance Elastography (MRE) and Diffusion Weighted Imaging (DWI), atomic force microscopy (AFM) and spinning disk confocal microscopy (SDCM).
This combination of optical and mechanical imaging techniques is a crucial step toward understanding the mechanical roles of cancer cells, stroma, and other non-cancerous components. How are the
bulk properties of tissues (MRE) influenced by the mechanical characteristics of subcompartments (AFM) that can be correlated with tissue geometry (SDCM) with subcellular resolution and in what
are the resulting changes in cancer progression?
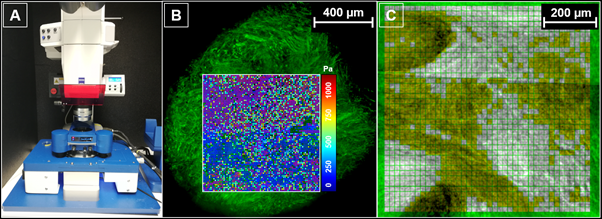 |
A: AFM setup, consisting of a Zeiss AxioZoom.V16 fluorescence microscope equipped with a JPK Hybridstage™.
B: Superposition of elasticity map and optical SDCM image, which shows the actin filaments in green color.
C: Using the optical image to create a mask for grouping and evaluating the mechanical data. |
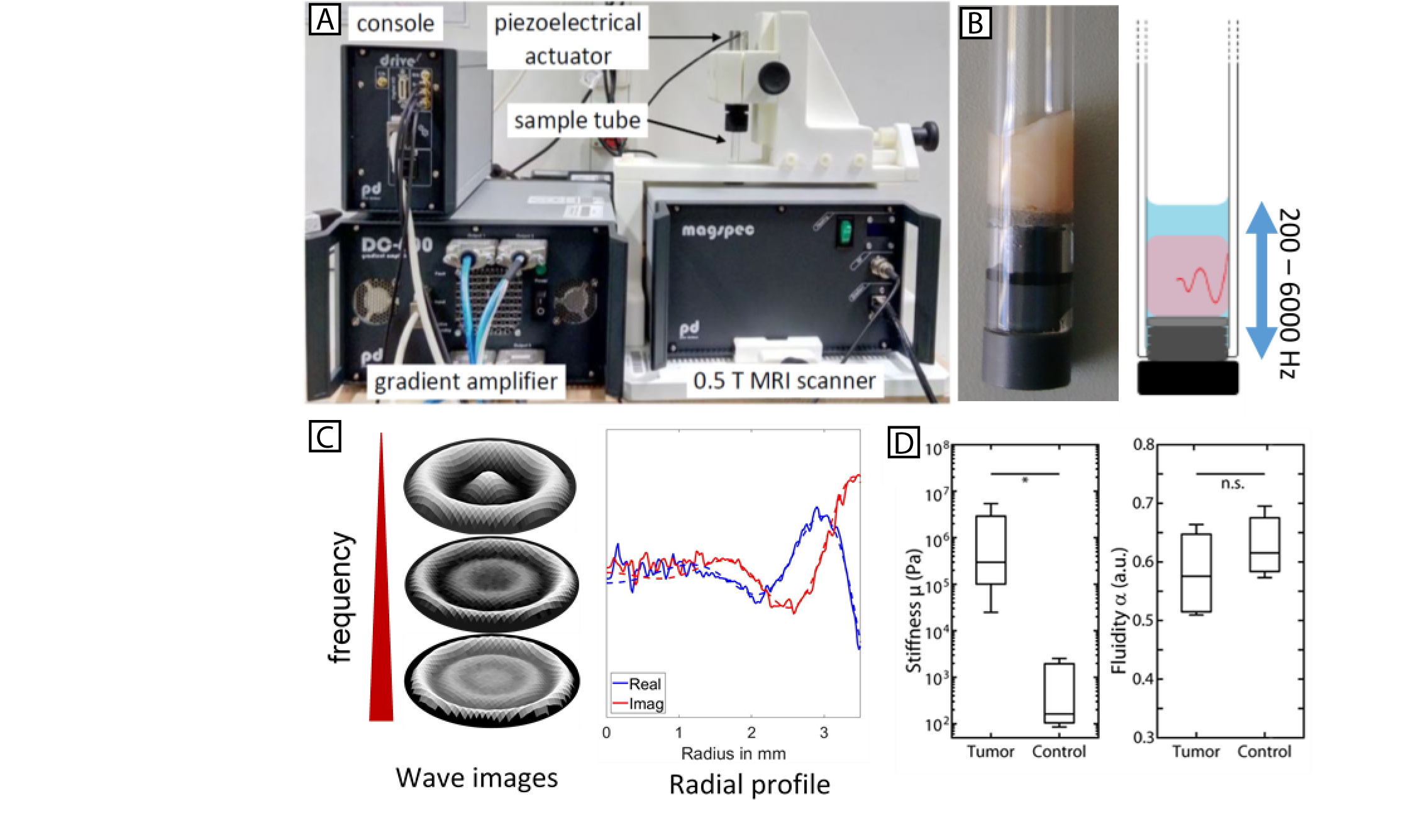 |
A: Tabletop-MRE system with 0.5 T MRI scanner, piezoelectric actuator with mounted sample tube, gradient amplifier
and console with signal preamplifier.
B: 7 mm glass tube with tumor tissue sample (left) and schematic representation of sample excitation
and respective shear wave propagation.
C: Representative wave fields for different excitation frequencies and exemplary radial profile
for one frequency used for rheological analysis.
D: Stiffness and Fluidity of primary human breast tumor vs. respective control tissue. |
Main techniques:
- atomic force microscopy
- MRE/DWI
- SDCM
Collaborators:
Prof. Ingolf Sack, Charité Berlin
Prof. Bahriye Aktas, Department of Gynecology, Women's and Children's Centre, University Hospital Leipzig
Tumor invasion and the role of the tumor microenvironment
A key step during cancer progression is the formation of metastases. Therefore single cells and cell clusters detach from the primary tumor and migrate through the body.
During this process the cancer cells need to adapt to different conditions and environments, such as collagen rich ECM or fatty tissue.
Our research focuses on the biomechanical properties that need to change when cells migrate in these different environments. Together with our collaborators from the
University Hospital Leipzig, we are also looking at the biomolecular crosstalk that drives these mechanical changes to get a detailed understanding.
Furthermore we are investigating the physics of cancer cell invasion into other tissues and trying to determine the role of the ECM in this process.
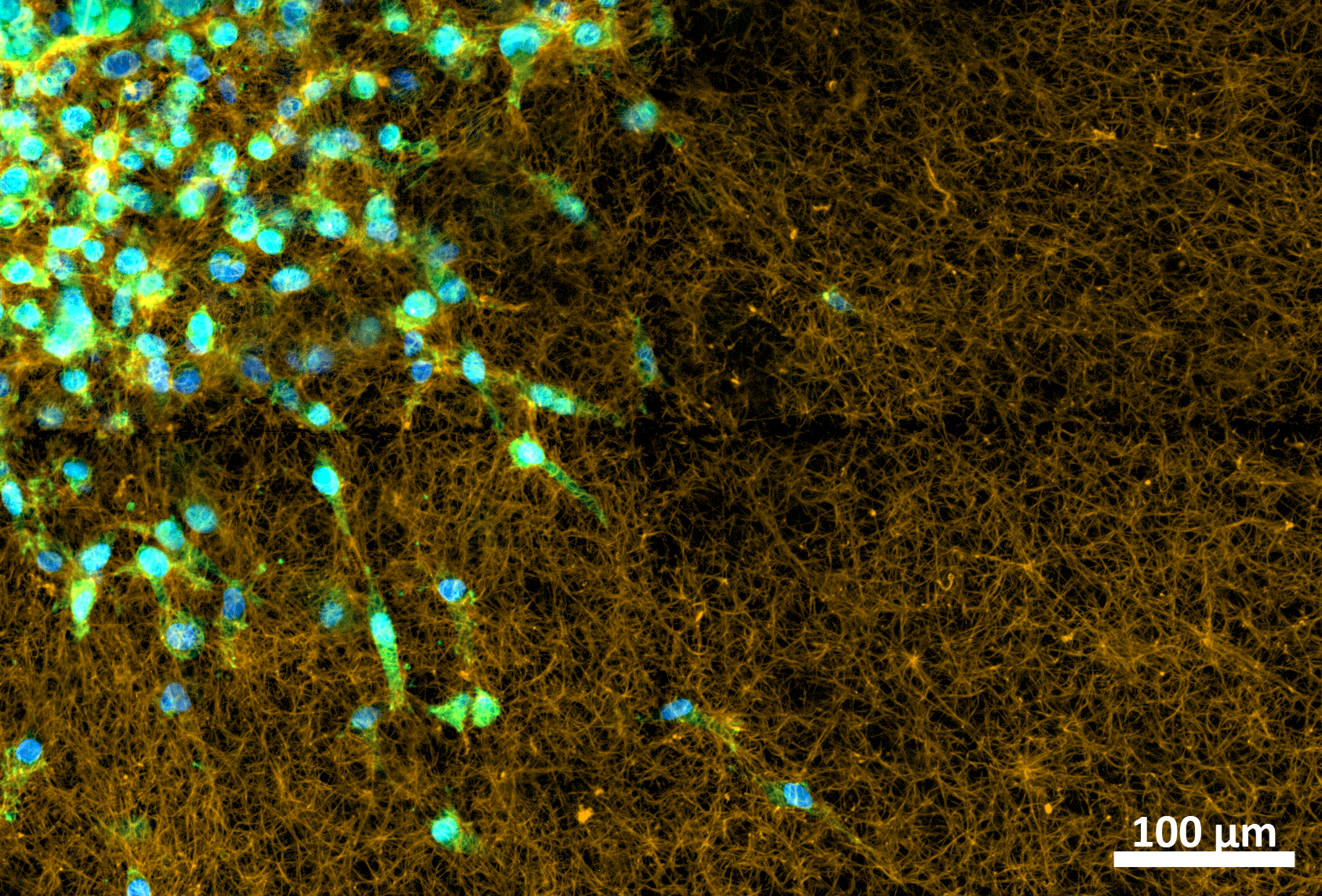
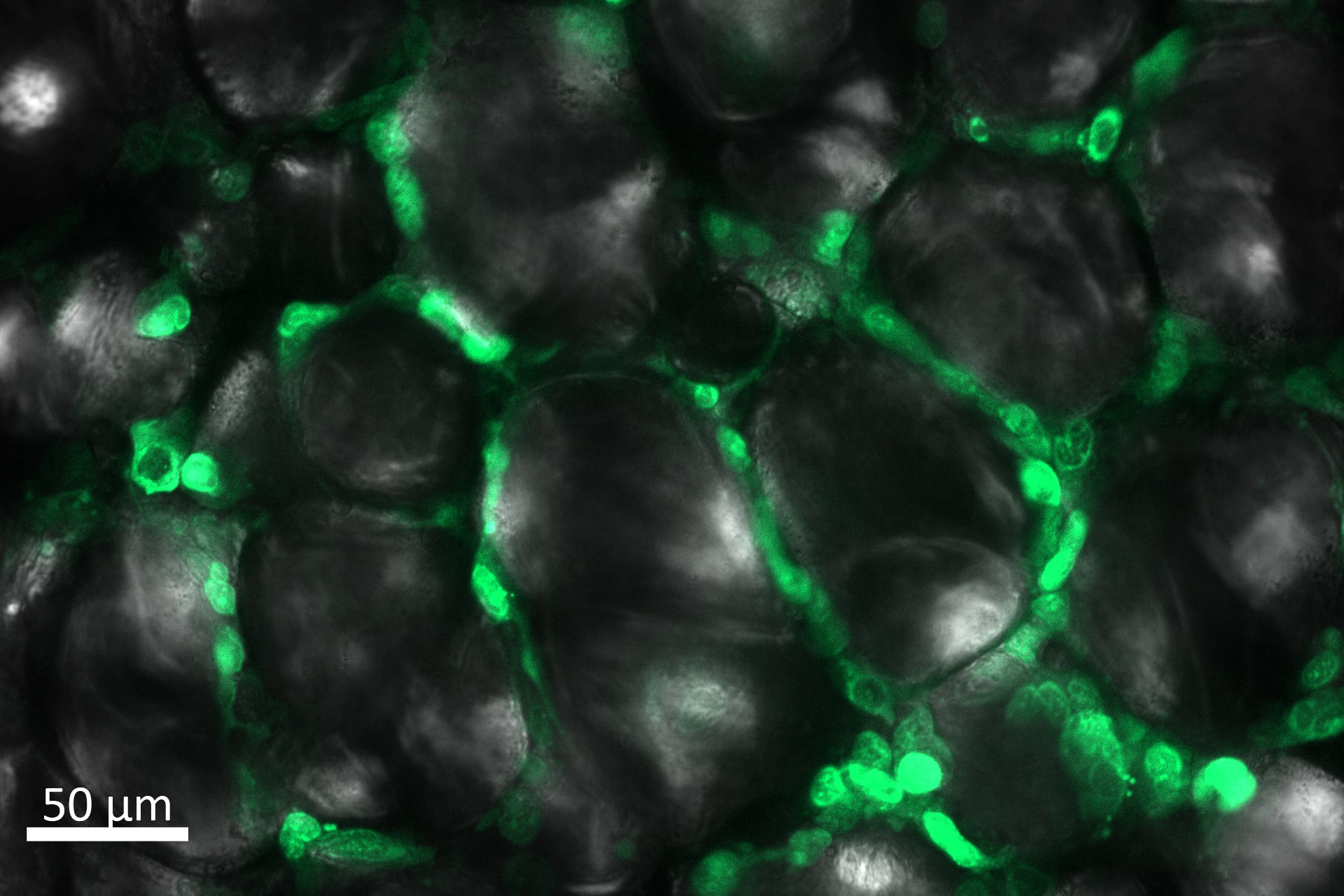 |
| MDA-MB-231GFP cells invading into different microenvironments. Left: extracellular matrix, Right: primary adipose tissue. |
Main techniques:
- live cell imaging
- 3D cell culture
- 3D cell tracking
- optical stretcher
Key publication:
Collaborators:
Prof. Matthias Blüher, Prof. Peter Kovacs, Dr. Dorit John - IFB Adiposity Diseases University Hospital Leipzig
Prof. Bahriye Aktas, Department of Gynecology, Women's and Children's Centre, University Hospital Leipzig
Dr. Jürgen Schiller, Dr. Jenny Leopold - Institute for Medical and Biophysics, University of Leipzig
Cancer cell motility through confinements
The extra-cellular microenvironment has a fundamental role in tumour growth and progression, and strongly affects the migration strategies adopted by single cancer cells during metastatic invasion.
The research carries out in the Käs Lab focuses on the ability of mesenchymal and epithelial breast carcinoma cells to migrate through three-dimensional narrowing microstructures upon chemoattractant
stimulation, and addresses the question of how the dynamical restructuring of the filamentous actin cytoskeleton modulates cell migration through the constrictions.
Additionally, we investigate possible correlations between the migration behavior in the microstructures and the expression of vimentin and keratin intermediate filaments in our cancer cell lines.
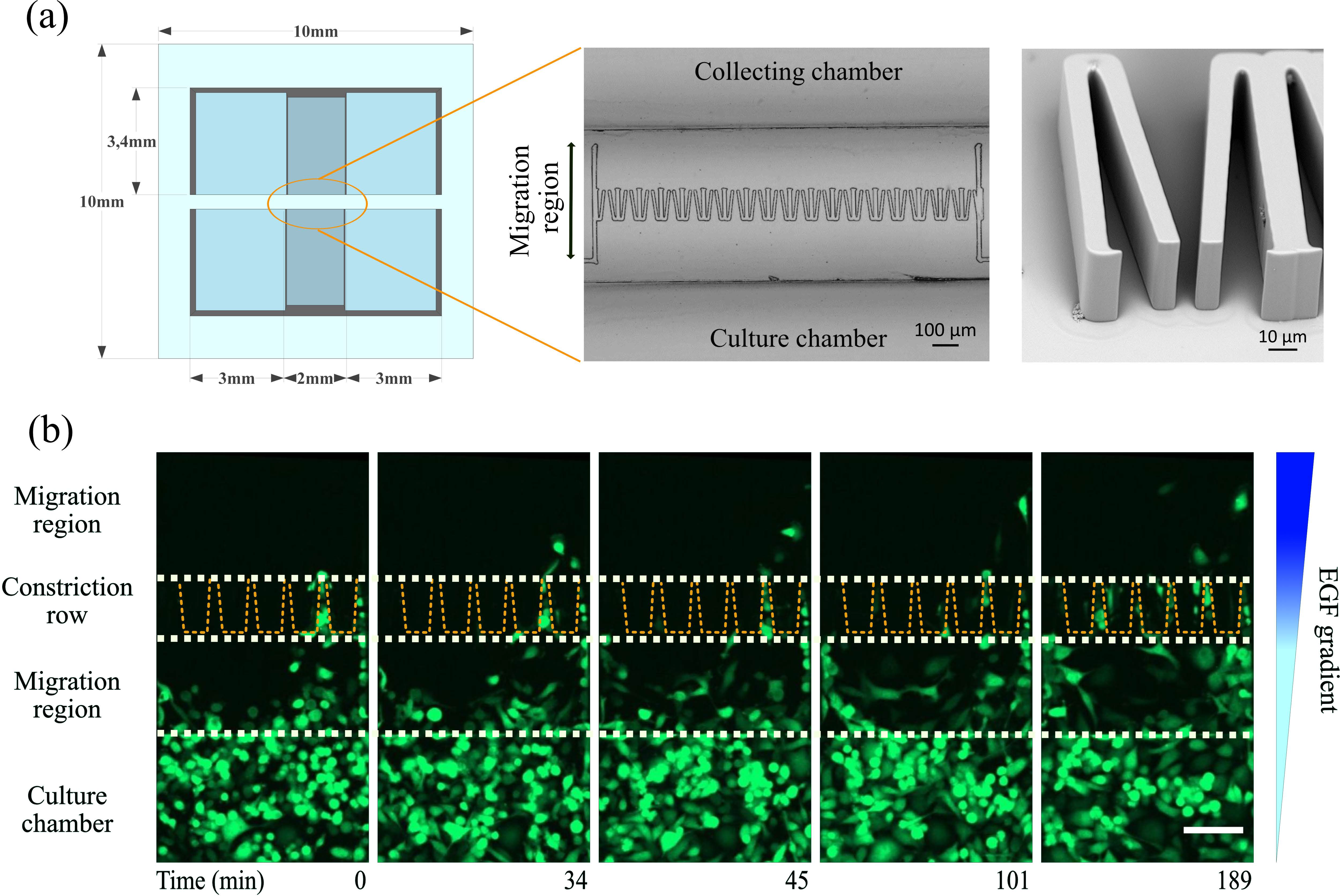
| Overview of the micro-constriction device:
(a) The device consists of two open reservoirs, separated by a row of funnel-shaped micro-constrictions which is covered by a 2 mm thick silica glass. The right panel shows a zoomed view of the micro-funnel row by scanning electron microscopy.
(b) MDA-MB-231 carcinoma cells with stable cytosol expressing GFP migrating through the constrictions. White dashed lines indicate the main sections of the migration region. Yellow dashed lines define the shape and location of the funnels.
Scale bar is 100 μm. The image is taken from Ficorella et al. 2019. |
Main techniques:
- 2D cell culture
- live fluorescence staining and immunostaining
- phase contrast and fluorescence microscopy
Key publications:
C. Ficorella et al, Normal epithelial and triple-negative breast cancer cells show the same invasion potential in rigid spatial confinement New Journal of Physics (2019)
DOI: 10.1088/1367-2630/ab3572
F. Sala et al, Rapid Prototyping of 3D Biochips for Cell Motility Studies Using Two-Photon Polymerization Front. Bioeng. Biotechnol. (2021)
DOI: 10.3390/bios12080604
C. Ficorella et al, Intermediate filaments ensure resiliency of single carcinoma cells, while active contractility of the actin cortex determines their invasive potential New Journal of Physics (2021)
DOI: 10.1088/1367-2630/ac1899
Collaborator:
Prof. Roberto Osellame, Politecnico di Milano, Institute for Photonics and Nanotechnologies, Italy
The role of molecular signatures in cancer prognosis
Molecular signaturer are sets of genes utilized for predicting the outcome such as death of a patient. Several signatures have been clinically validated and are used in clinics.
However, It is not yet clear what informational gain do these signatures procure and if their prognostic power is fundamentally limited.
We investigate the utility and extensibility of molecular signatures by asking following questions:
- What informational gain do molecular signatures procure?
- Does a fundamental maximum prognostic limit of molecular signatures exist?
- How can one incorporate biophysics into the development of molecular signatures?
Main techniques:
- Database analysis
- Statistical analysis of gene expressions
- Machine learning
Key publication:
D. Tschodu et al, Comparative analysis of molecular signatures reveals a hybrid approach in breast cancer: Combining the Nottingham Prognostic Index with gene expressions into a hybrid signature PLoS One (2022)
DOI: 10.1371/journal.pone.0261035
Collaborator:
Prof. Axel Niendorf, Pathologie Hamburg-West, Institut für Histologie, Zytologie und Molekulare Diagnostik
Genomics and Bioinformatics Research in Cancer
Genomics research is the study of the complete set of DNA (including all of its genes) and DNA -related structures such as; gene expression, methylation, mutation, microRNA and alteration of
their expression and structures in a person or other organism.
Genomics researchers analyze enormous amounts of DNA-sequence data to find variations that affect health, disease or drug response by using high-performance computing and math techniques known as
bioinformatics and biostatistics tools and databases. In humans that means searching through about 3 billion units of DNA across 23,000 genes. As a result of genomics research, it is possible to
predict, diagnose, and treat diseases more precisely and personally than ever.
To analyze this kind of big data, different machine learning techniques and statistical approaches are needed. Not only the data which are produced in our lab, but also publicly open access
genomics databases are used to compare our laboratory results simultaneously.
Key publications:
S. Dogan et al, Structural Analysis of microRNAs in Myeloid Cancer Reveals Consensus Motifs Genes (2022)
DOI: 10.3390/genes13071152
H. Herzog et al, Targeted Sequencing of Plasma-Derived vs. Urinary cfDNA from Patients with Triple-Negative Breast Cancer Cancers (2022)
DOI: 10.3390/cancers14174101
Collaborators:
Prof. Bahriye Aktas, Dr. Ivonne Nel, Department of Gynecology, Women's and Children's Centre, University Hospital Leipzig, Germany
Prof. Abdulhamit Subasi, Turun yliopisto - University of Turku, Faculty of Medicine, Finland
Emrulla Spahiu, Institute of Molecular and Cell Physiology, Hannover Medical School, Germany
Anis Cilic, Excellence Cluster Cardiopulmonary System, University of Giessen and Marburg Lung Center (UGMLC), Justus-Liebig-University
Prof. Tuncay Delibasi, University of Texas Southwestern Medical Center, USA
|
|













