Objectives: To understand the molecular basis of mitochondrial function, the crosstalk between nucleus and mitochondria as well as the fate of mitochondria during embryogenesis, tissue development and disease.
Approach: We use a number of different approaches to study the biogenesis of mitochondria: a) development of a DNA vector that allows the destruction of endogenous mtDNA; b) development of a mitochondrial fluorescently activatable GFP (mtFAGFP) to study fission and fusion of mitochondria in vivo; c) development of transport vectors for trafficking nucleic acids to mitochondria, d) characterization of mtDNA isolated from tumor tissues, e) tagging HREs and other mito-nuclear factors with FAGFP to study their cellular fate by live imaging technologies.
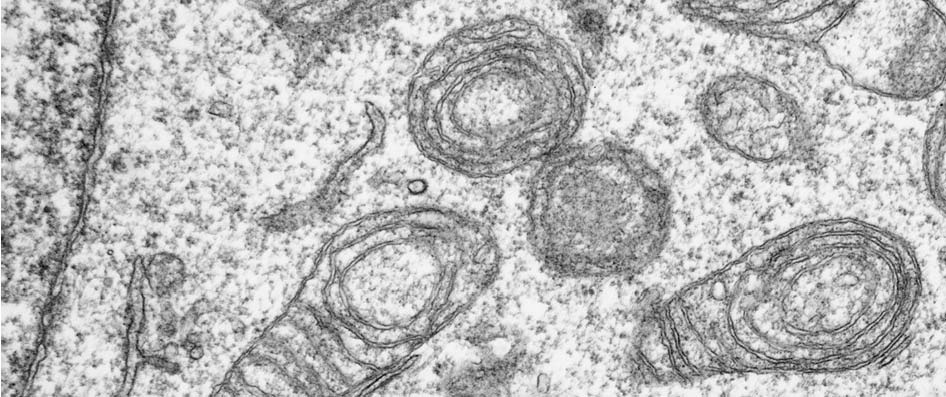
Progress: Since our start at the Biocenter some years ago, we were able to generate a cell line devoid of endogenous mtDNA by treating the parental cells with low dosages of ethidium bromide. The cell line provides a nuclear marker (BrdU) that can be used in subsequent cytoplast fusion experiments to transfer exogenous mitochondria to our recipient and thus to study their behavior on a different nuclear background. We found that mitochondria devoid of any endogenous mtDNA display an awkward membrane structure that can be described as an onion-shaped membrane system (see figure 1). Although similar mitochondria have been observed in patients with mitochondrial disorders, the molecular mechanism generating these structures remains unclear.
To overcome the mutagenic effects of ethidium bromide, we constructed a vector system that leads to the expression of mitochondria targeted restriction enzymes. Utilizing the vector, we can destroy endogenous mtDNA by an enzymatical process rather than toxifying cells with mutagens such as ethidium bromide. Further on, we developed a photo activatable GFP derivative that includes a mitochondrial signal sequence. This tool will be used in studying fusion and fission events of mitochondria. To relocate nucleic acids to mitochondria, we developed a peptide DNA construct. The construct consists of a mitochondrial signal peptide and a DNA moiety so that translocation of nucleic acids to mitochondria can be achieved.
Significance: Gene defects in the mitochondrial genome lead to a variety of devastating human diseases. While genetic information on the diseases is readily available, very little is known about the pathogenic processes that are caused by the mutations. Moreover, it seems to date that the nuclear driven biogenesis of these organelles, including fusion and fission events of mitochondria, can modulate the expression of pathogenic or deleterious mtDNA variations. By studying mitochondrial biogenesis, we expect to gain insight into the relevant pathogenic events in patients presenting with mitochondrial disorders.
Future projects: Our plan is to use transgenic approaches to follow the fate of mitochondria during development, embryogenesis and disease. For this purpose, we want to establish cell lines that express the mitochondria targeted restriction endonuclease upon gene induction. These cells can be used to generate a variety of rho zero cells (devoid of mtDNA) so that mitochondrial-nuclear crosstalk can be examined. Further on, we want to generate mitoGFP transgenic animals to follow the fate of intact organelles in different tissues. This approach will be of special interest to clinical phenotypes, since most mitochondrial diseases express defects with high tissue specificity.
Collaborations: Prof. Dr. Gaetano Villani (University of Bari, Italy), Prof. Dr. Antonio Moschetta (Consorzio Mario Negri Sud, Chieti, Italy), Prof. Dr. Ian Trounce (University of Melbourne, Australia), Prof. Dr. Douglas C. Wallace (University of California, Irvine, USA), Prof. Dr. Robert N. Lightowlers (University of Newcastle, UK), Prof. Dr. Constantine Sekeris (University of Athens, Greece), Prof. Dr. Klaus Scheller (Biozentrum, Würzburg, Germany), Prof. Dr. Georg Krohne (Biozentrum, Würzburg, Germany)