Introduction Actin
Dynamics and Kinetics Actin
Dynamics and Kinetics
|
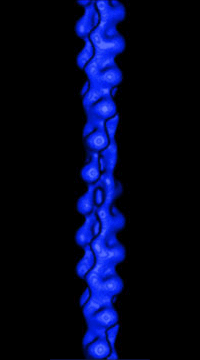 |
|
(Figure from Julie Hodgkinson, Imperial College School
of Medicine, National Heart and Lung Institute) |
|
|
Actin is a semi-flexible polymer found as a part of the cytoskeleton of
cells in higher biological organisms. Its semi-flexible nature means that
the typical lengths of the filaments in solution are on the order of its
persistence length, LP, or the distance over which the angles
of the ends become uncorrelated. In simple terms, this means the persistence
length represents the distance over which bending at one end does not affect
the other end. In vitro, persistence length of actin filaments is
on the order of 15 µm.
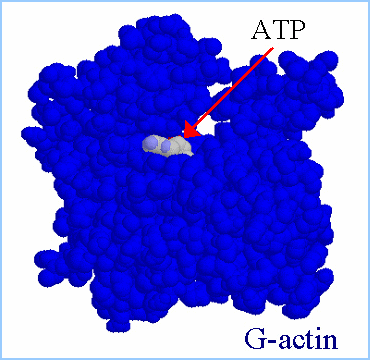
Actin filaments consist of repeating subunits of actin monomers in a
right-handed double helical structure. The individual units or G-actin
(globular actin) are 43 kD in size, and have a diameter of approximately
5 nm. The monomers have several subunits, and an internal cleft which can
bind ATP and Magnesium ions. Each monomer has an asymmetrical structure,
giving polymerized actin filaments a "polarity", with the ends referred
to as the "pointed" (or minus) and "barbed" (or plus) ends.
Actin monomers move freely throughout solution according to Brownian
diffusion. At a critical concentration of 0.1 mg/ml, the monomers begin
to form stable nuclei consisting of 3-4 individual units. At this point,
the filaments begin their elongation phase, where free monomers add to
both sides of the filament, hydrolyzing ATP and releasing inorganic phosphate
in the process. While filaments add to both sides of the filaments, they
do not add with equal rates – the barbed, or plus end is the fast-growing
end, while the pointed, or minus end is the slow-growing end.
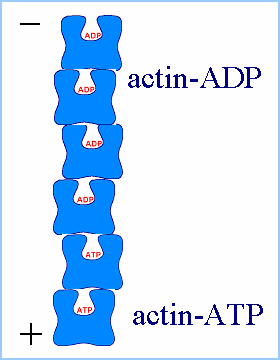 |
|
| (Figures from Joyce J. Diwan,
Rensselaer Polytechnic Institute) |
|
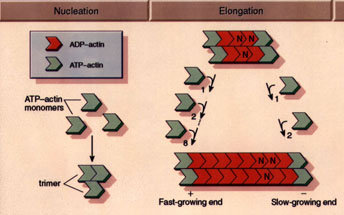 |
|
(Figure from Professor Daniel L. Purich, Department
of Biochemistry & Molecular Biology, University of Florida College
of Medicine) |
The polymerization rate depends largely on the concentration of the
monomer pool, so a balance is eventually reached between these two factors,
and the filaments enter a steady-state phase. In this state, the filaments
undergo a process referred to as "treadmilling", whereby there is a constant
polymerization and depolymerization of the filaments. On average, the filaments
polymerize at their barbed ends and depolymerize at their pointed ends,
so there is a net transport of monomers from the barbed to pointed ends.
This process is a non-equilibrium effect as ATP is being constantly hydrolyzed
and the system continually remains in a dissipative state.
|
 |
|
(Figure from Josef A. Käs, University of Leipzig) |
Individual actin filaments are only approximately 6 nm in width, several
orders of magnitude less than the minimal resolution of a light microscope
(~200 nm). As a result, ordinary light microscopy methods are not used
for visualization of actomyosin networks in solution. Techniques previously
developed to visualize single actin filaments through fluorescent microscopy
in F-actin networks (Käs, 1994) are frequently utilized to microscopically
investigate actin-myosin solutions. This involves labeling each actin monomer
with a rhodamine molecule bound to an antibody raised against rabbit skeletal
muscle actin. The rhodamine is excited by green light and fluoresces brightly
red. In this way, each monomer on the actin filament (approximately 4000
monomers per 10 µm filament) acts as a pinhole emitting light, thus
making individual filaments visible through fluorescent microscopy.
Pattern Formation of Actin Filaments
  Actin
filaments pose an interesting and difficult challenge for contemporary
polymer physics. They are semi-flexible, with a bending stiffness much
greater than that of flexible biopolymers such as DNA, but significantly
more flexible than rigid rod-like macromolecules such as Microtubules.
Being relatively rigid, actin polymers can undergo strong steric interactions
with one another, and concentration -dependent phase transitions occur
in which the system partially enters a liquid crystalline phase. We have
seen evidence of an even higher degree of ordering characterized by large
scale spatial structuring. We are currently using a variety of microscopy
techniques, including fluorescence, confocal, laser scanning microscopy
and a novel polarization device to investigate the appearance of a concentration-
dependent order parameter in the system. Our aim is to characterize both
patterns and the parameter space in which they appear. Actin
filaments pose an interesting and difficult challenge for contemporary
polymer physics. They are semi-flexible, with a bending stiffness much
greater than that of flexible biopolymers such as DNA, but significantly
more flexible than rigid rod-like macromolecules such as Microtubules.
Being relatively rigid, actin polymers can undergo strong steric interactions
with one another, and concentration -dependent phase transitions occur
in which the system partially enters a liquid crystalline phase. We have
seen evidence of an even higher degree of ordering characterized by large
scale spatial structuring. We are currently using a variety of microscopy
techniques, including fluorescence, confocal, laser scanning microscopy
and a novel polarization device to investigate the appearance of a concentration-
dependent order parameter in the system. Our aim is to characterize both
patterns and the parameter space in which they appear.
There is an additional interesting aspect to these actin systems known
as treadmilling. This is a process due to actin's unique reaction kinetics
whereby individual monomers are steadily cycled through a filament which
maintains an overall constant length. This is an energy dissipative process
and assures that the system cannot attain thermodynamic equilibrium. We
are currently investigating the role that this nonequilibrium process might
play in the formation of the spatial structures. Perhaps this will shed
some light on the role of treadmilling in cells, which as of yet remains
a mystery.
In the Presence of the Molecular Motor Myosin
II
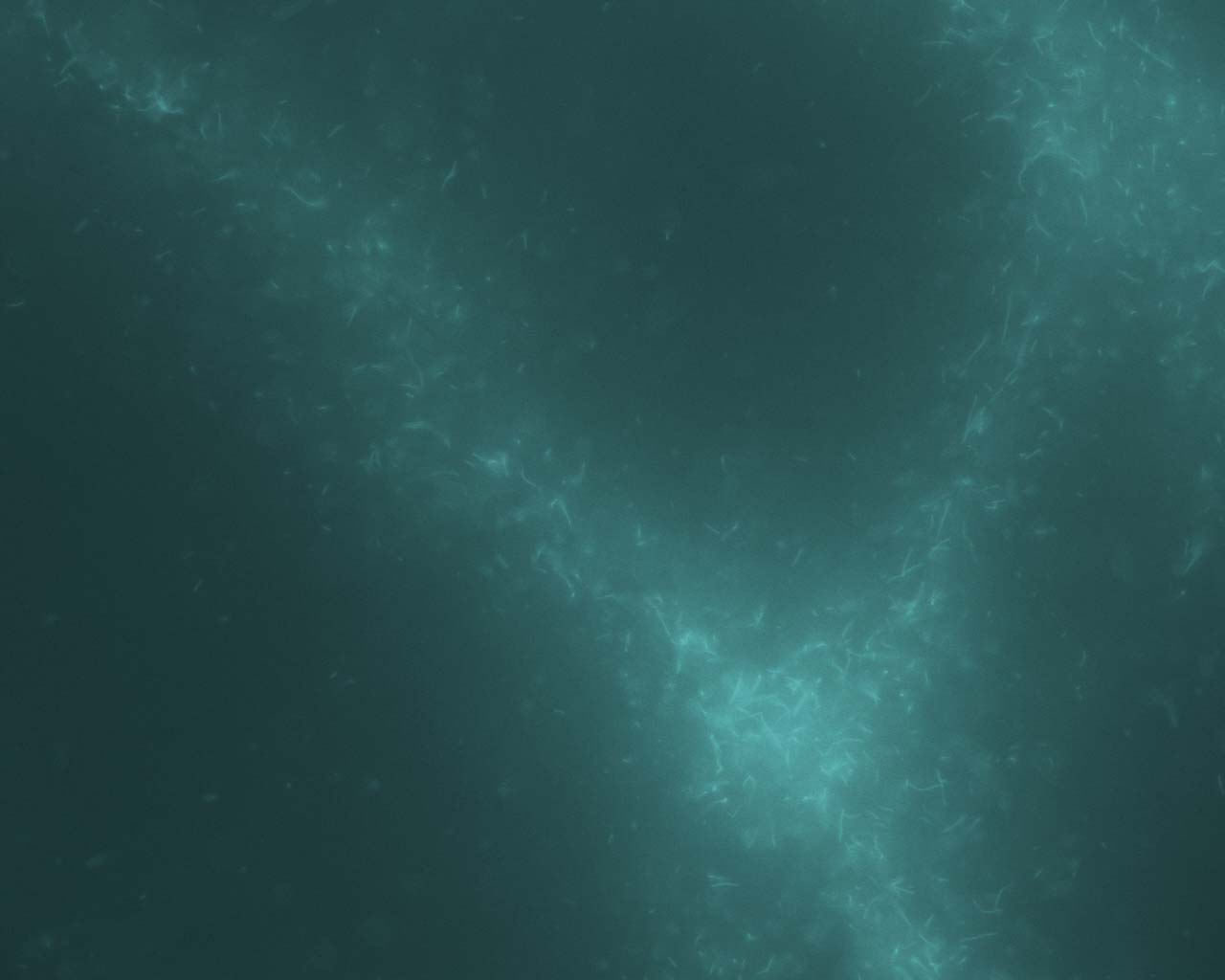 All
eukaryotic cells depend upon mechanisms of protein filament self-assembly
to form their cytoskeletons. The cell's need for motility and rapid response
to stimuli additionally require the existence of pathways which serve to
restructure and disassemble cytoskeletal elements. While temperature-driven
increases in disorder are the most physically fundamental methods for breaking
down complex structures, they would compromise the cell's viability. Molecular
machinery provides a non-destructive means to accomplish the same goals.
This method is utilized on the genetic level with the unfolding of DNA
strands for replication and cell division. Molecular machinery unfolds
the DNA strands without heat-induced damage to the cell, providing an alternative
to temperature-driven methods (Lodish et al, 2000). We have observed experimental
evidence of a similar mechanism functioning on actin cytoskeletal dynamics,
involving collections of the actin-specific molecular motor Myosin II.
Crosslink-driven bundling self-assembles complex actomyosin structures
in the near-chemical-equilibrium state, including bundles, asters, and
large aggregates. Activation of the motors, however, causes a rapid disassembly
of all structures. Such a mechanism is not only harmless to cell function,
but occurs on a very rapid timescale which is favorable for quick cytoskeletal
dynamics. All
eukaryotic cells depend upon mechanisms of protein filament self-assembly
to form their cytoskeletons. The cell's need for motility and rapid response
to stimuli additionally require the existence of pathways which serve to
restructure and disassemble cytoskeletal elements. While temperature-driven
increases in disorder are the most physically fundamental methods for breaking
down complex structures, they would compromise the cell's viability. Molecular
machinery provides a non-destructive means to accomplish the same goals.
This method is utilized on the genetic level with the unfolding of DNA
strands for replication and cell division. Molecular machinery unfolds
the DNA strands without heat-induced damage to the cell, providing an alternative
to temperature-driven methods (Lodish et al, 2000). We have observed experimental
evidence of a similar mechanism functioning on actin cytoskeletal dynamics,
involving collections of the actin-specific molecular motor Myosin II.
Crosslink-driven bundling self-assembles complex actomyosin structures
in the near-chemical-equilibrium state, including bundles, asters, and
large aggregates. Activation of the motors, however, causes a rapid disassembly
of all structures. Such a mechanism is not only harmless to cell function,
but occurs on a very rapid timescale which is favorable for quick cytoskeletal
dynamics.
Other Actin Dynamics Experiments
We are investigating the effect of cytoplasm contents on the bending
stiffness of individual filaments. From these experiments we hope to gain
a more accurate picture of the stiffness of actin filaments in vivo, as
this property is crucial to the integrity of the actin cytoskeleton. We
are also using torsion pendulum rheometry to explore bulk properties of
actin networks under a variety of conditions, including in the presence
of cross-linking proteins. We hope that these in vitro investigations will
shed new light on the dynamic properties of the actin cytoskeleton.
|










 Actin
filaments pose an interesting and difficult challenge for contemporary
polymer physics. They are semi-flexible, with a bending stiffness much
greater than that of flexible biopolymers such as DNA, but significantly
more flexible than rigid rod-like macromolecules such as Microtubules.
Being relatively rigid, actin polymers can undergo strong steric interactions
with one another, and concentration -dependent phase transitions occur
in which the system partially enters a liquid crystalline phase. We have
seen evidence of an even higher degree of ordering characterized by large
scale spatial structuring. We are currently using a variety of microscopy
techniques, including fluorescence, confocal, laser scanning microscopy
and a novel polarization device to investigate the appearance of a concentration-
dependent order parameter in the system. Our aim is to characterize both
patterns and the parameter space in which they appear.
Actin
filaments pose an interesting and difficult challenge for contemporary
polymer physics. They are semi-flexible, with a bending stiffness much
greater than that of flexible biopolymers such as DNA, but significantly
more flexible than rigid rod-like macromolecules such as Microtubules.
Being relatively rigid, actin polymers can undergo strong steric interactions
with one another, and concentration -dependent phase transitions occur
in which the system partially enters a liquid crystalline phase. We have
seen evidence of an even higher degree of ordering characterized by large
scale spatial structuring. We are currently using a variety of microscopy
techniques, including fluorescence, confocal, laser scanning microscopy
and a novel polarization device to investigate the appearance of a concentration-
dependent order parameter in the system. Our aim is to characterize both
patterns and the parameter space in which they appear.
